Abstract
Peptides can self-assemble themselves and create various assemblies with a variety of structural and catalytic properties. In this project, I will investigate the interactions between heterogenous amyloids created by the Ac-IHIHIGI-NH2 peptide with zinc. This information is useful for the creation of synthetic analogues of natural enzymes in biotechnological settings.
Introduction
Metals are significant in enzyme catalysis, in this project, the interactions of heterogenous amyloids that are formed by Ac-IHIHIGI-NH2 when combined with zinc will be analyzed. Such data will be used to understand how the peptide will interact with environmentally and biologically relevant molecules.
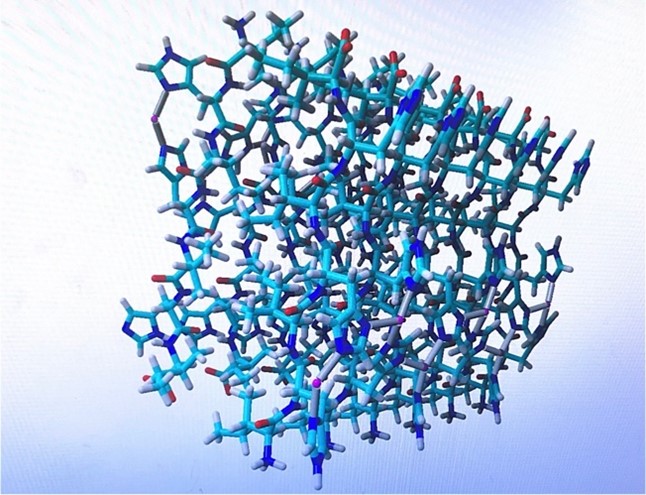
Figure 1. Peptide on YASARA Software with eight zincs on the structure
Experimental Details
Using computational techniques, the molecular modeling of the interactions between zinc and Ac-IHIHIGI-NH2 will be processed. First, the ligand, Nitro Phenyl Phosphate (NPP) will be created in Gaussian View. Then, the peptide will be created on YASARA while differentiating between which molecules will need to use the amino acid HIS or HIX based on whether they are single bonds or double bonds. After, the substrates are docked on the amyloid using Auto Dock (version 4.0). The lowest energy conformer will be taken from the docked structure to perform molecular dynamics stimulations with the amyloid-substrate placed in an aqueous solution for the scene. All MD stimulations will be done through “GROMACS” and “YASARA” software’s. During the molecular dynamic’s performances, the starting molecules will have their energy minimized for 2000 steps. First, topology folders are created and after a cubic box is made around the subject. Then water is added into the box to make it the aqueous solution. Then energy minimization will be prepared and run for 2000. steps are run. Afterwards, it is submitted for npt equilibration and for md calculation. The results of these minimizations will create the initial structure for the MD stimulations. The MD stimulations will produce RSMD files with protein after lsq fit to backbone and backbone after lsq fit to Backbone, also an RMSD of protein after lsq fit to protein. The files will display the significant changes in protein structures compared to the initial starting point.
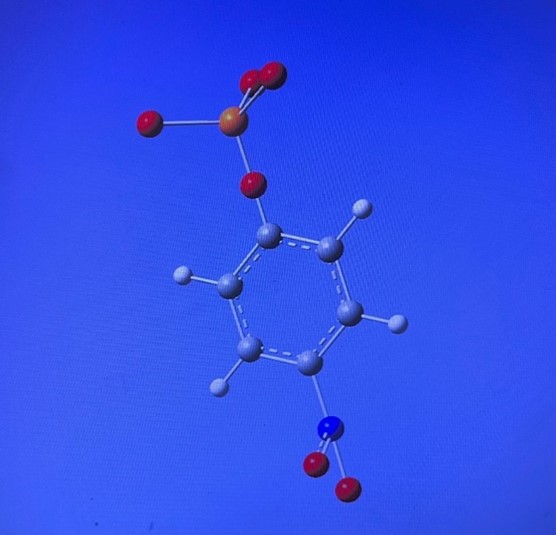
Figure 2. NPP structure designed to create the ligand containing benzene ring in the center
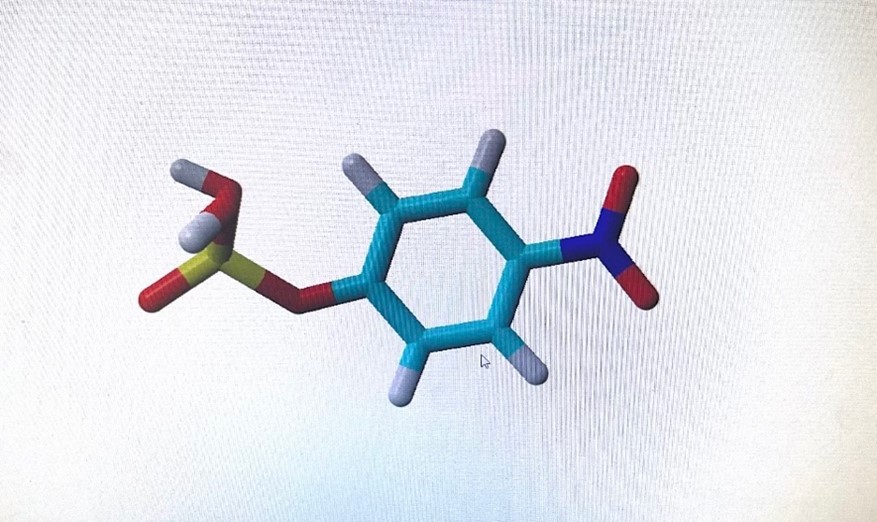
Figure 3. Ligand structure on YASARA after being designed and stimulated
Results and Discussions
The molecular dynamics stimulations displayed the metal, zinc, creating a distance from the parallel peptide that was assembled with the amino acid histidine and others. The hydrophobic interface was stabilized by a polar chain, and the hydrated interface maintained the zincs binding to the amino acid histidine. A final histidine completed the protein ligand site and left a free site on the zinc. Zinc presents to be an advantageous metal to determine the structures of metal amyloids. The amyloids provide insight on the functions of natural enzymes which is beneficial in designing their synthetic analogues.
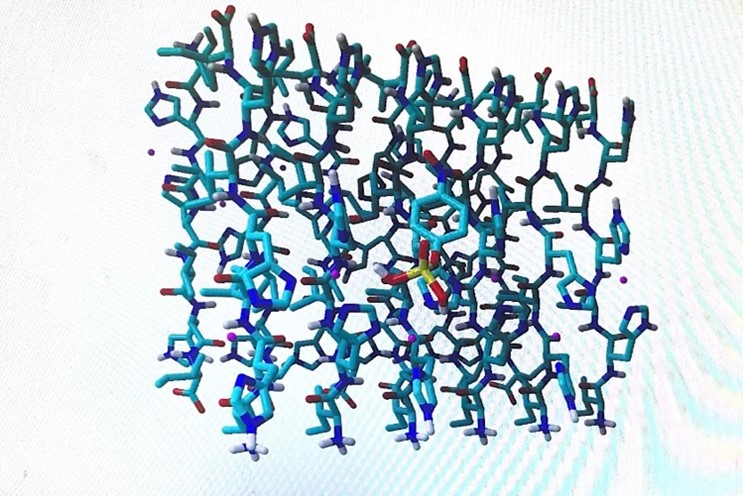
Figure 4. Peptide with ligand after docking was completed through AutoDock version 4.0
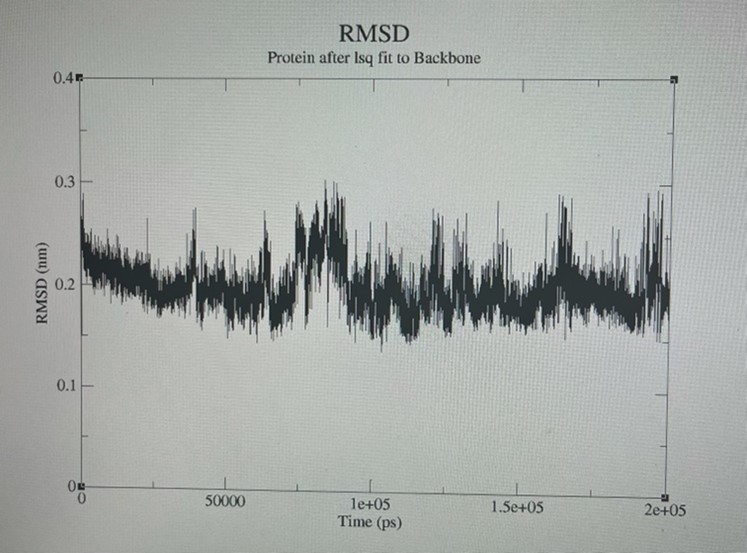
Figure 5. Root means square deviation (RMSD) of the protein fit to the backbone group derived from MD trajectories.
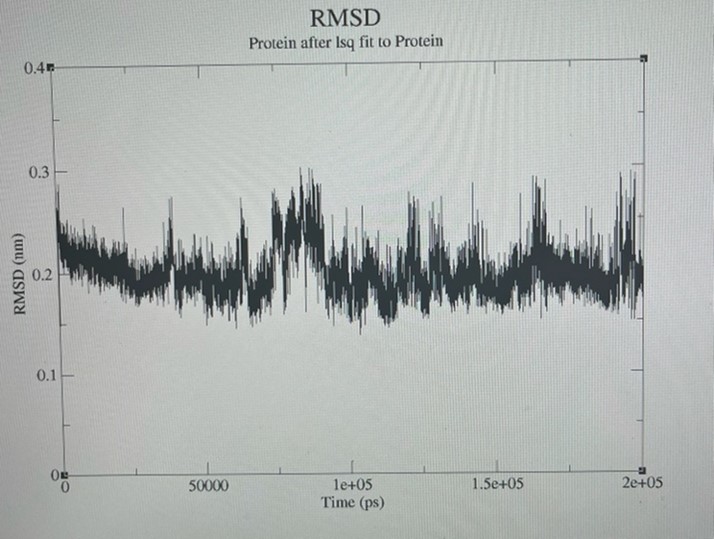
Figure 6. Root means square deviation (RMSD) of the protein fit to the protein group derived from MD trajectories.
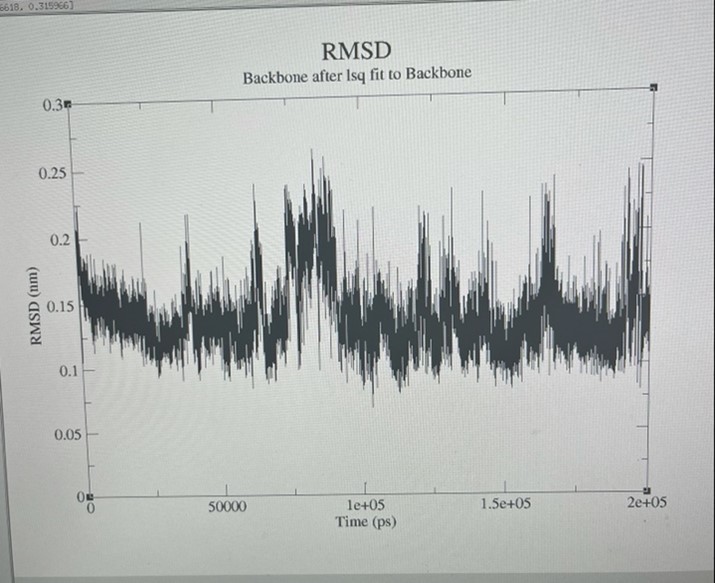
Figure 7. Root means square deviation (RMSD) of the backbone fit to the backbone group derived from MD trajectories.
Conclusion
Amyloids have recently been used to catalyze hydrolysis and oxidation reactions. They are key structures in both proteins and pathologic proteins misfolding. When the beta- strands of an amyloid structure are parallel; side chains of each position align which makes for good ligands that attach to the zinc. An intricate interplay by which metal ions stabilize amyloid structures is revealed from the stimulations which then shapes ligand geometry and the catalytic reactivity of zinc.
Acknowledgements
I would like to acknowledge the Chemistry Department of the University of Miami through their partnership with the American Chemical Society for the opportunity to conduct such computational research in their laboratory. I would also like to acknowledge the American Chemical Society and their Project Seed program that has allowed me to advance my understanding of the world around me. Finally, I would like to thank Professor Rajeev Prabhakar for explaining the processes that needed to take place to conduct the experiment.
References
1. Myungwoon Lee, Tuo Wang, Olga V. Makhlyne, Yibing Wu, Nicholas F. Polizzi, Haifan Wu, Pallavi M. Gosavi, Jan Stöhr, Ivan V. Korendovych, William F. DeGrado and Mei Hong (2017) Zinc-binding structure of a catalytic amyloid from solid-state NMR.
2. Liam R Marshall, Megha Jayachandran, Zsofia Lengyel-Zhand, Caroline M Rufo, Austin Kriews, Min-Chul Kim, Ivan V Korendovych (2020) Synergistic Interactions Are Prevalent in Catalytic Amyloids










