Abstract:
Peptides are precursors of proteins and oxygen and are an encouraging component for creating adaptable, self-healing molecular contracts. Recent developments in the field give information about stable and self-healing peptides that are remarkably active at stimulating a variety of processes. Through the uses of molecular dynamics we are able to understand the structural and catalytic properties of peptides with various amino acid.
Introduction:
Pharmaceuticals drive for the discovery of catalyst to enhance and improve the effectiveness of already existing drugs. This is done so through the modification of peptides that can make multiple assemblies with various catalytic properties. This self-assembly is a process in which there are spontaneously formed ordered clusters without any need of human interaction. Peptides offer much opportunity for self-assembly due to their ability to interact through Van der Waals forces, hydrogen bonding, electrostatic, and stacking interactions to form various structures that can differ in hydrophobicity, charge, and size. This allows for researchers to manipulate peptides to form efficient hydrogels. These hydrogels can be used for wound healing, catalysis, study of Aggregation, etc. Additionally, through the use of artificial amino acids there is a wide verity of modifications that can be made to the architectural structure of the peptide. This is important as the use of an artificial amino acids can result in better activity, allows for different types of stacking interactions, and allows for the use of other metals which increase the scope of the catalysis.
In this project, we used a artificial 9 peptide chain with a synthetic amino acid (ALF) which has very similar properties to that of Histidine in hopes of increasing activity. The ALF residue on bordering peptides was placed to form an active site to bond with the cofactor Zn. Zinc is present in metalloenzymes of carbonic anhydrase that can catalyze a large number of reactions. The two artificial 9 peptide chains (ALF-LQLQLQ-ALF- NH2 & Ac-ALF-LRLRLRL-ALF-NH2) consisted of the natural amino acids Glutamine, Leucine, and Arginine. Glutamine has polar uncharged side chains that allows for the creation of hydrogen bonds between the amino acid chains to help maintain its structure. Leucine has a hydrophobic side chain which allows for the creation of a hydrophobic core to prevent the non-hydrophobic amino acids from creating hydrogen bonds with water molecules. And the Arginine is the charged amino acid that causes the self-healing property forthe peptide chain.
We will use software such as GROMACS and YASARA to give us accurate molecules when put under certain conditions. The molecules created will be put through molecular dynamic simulations which resolve atomic movement equations to compute the equilibrium and transport properties of the systems. This is especially useful in medicinal research as it is used for the prediction of protein-ligand docking. The purpose of molecular dynamics in this research is to study physical interactions within the molecule under standard temperature and pressure (293.15K & 14.696 psi) which will result in an equilibrated structure.
Computational Details:
Through the use of software, we were able to compute physical interactions between the molecules. We used the GROMACS 2022 program to run all of our molecular dynamics simulations (MD) which put the molecule under SPT and form an equilibrated structure. The force field used for the MD was AMBER 99sb which was used to reproduce the molecular geometry of the structure. For the 9-peptide chain we ran the MDs for 100 nanoseconds meaning the simulation will show the changes that that happen to the molecule during the first 100ns. For the molecule with the zinc, we used an already equilibrated structure, so we ran the MD for 50 nanoseconds. Since we used ALF which is an artificial amino acid, we had to create a set of parameters. First, we had to optimize the ALF structure in Gaussian 9 using 6 B3LYP functional with 6-31g(d) basis set. We then used this optimized AFL structure to calculate a set of parameters using ANTECHAMBER from gaussian output.
Results & Discussion:
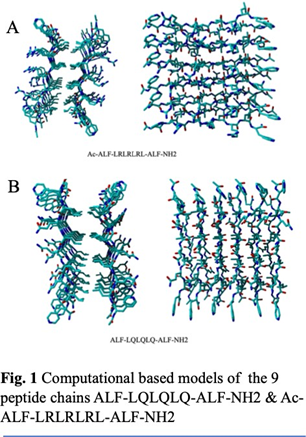
Through the use of YASARA, we were able to model and create peptide structures. For the first model we created a peptide chain using Leucine and Arginine (Fig. 1A). For the second model we used Leucine and Glutamine (Fig. 1B). We did this to create a hydrophobic core and to test catalytic activates. We maintained a alternating hydrophobic-hydrophilic sequence as they are reported to have amyloid like structures with good catalytic activity. The use of Arginine (R) allows for increased antimicrobial activity.
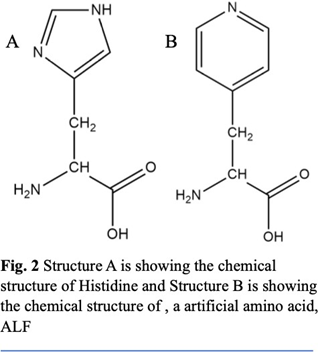
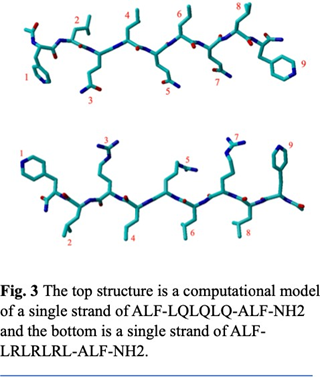
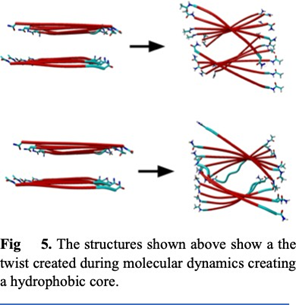
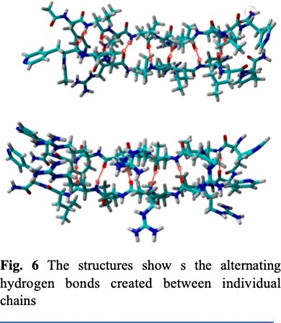
And metal binding residues like Histidine increases the aggregation. In both of the structures we used an artificial amino acid ALF instead of Histidine. ALF has a very similar structure to histidine (Fig. 2), allowing it to bond with metals due to the nitrogen on the para position in the benzene ring. We also used ALF as it has been previously reported to coordinate better than natural amino acids near neutral conditions. For this study we created two- different 9-peptide combinations for structural and activity studies (Ac-ALF-LQLQLQ-ALF- NH2 & Ac-ALF-LRLRLRL-ALF-NH2). As seen in fig. 3 points 1 and 9 are the active site ALF. Points 2, 4, 6, and 8 are the hydrophobic amino acids and points 3, 5, and 7 are the hydrophilic test sites. These structures were equilibrated for 100ns by Molecular Dynamics using Gromacs 2022 with AMBER99sb forcefield. After running molecular dynamics on the two structures Ac- ALF-LQLQLQ-ALF-NH2 & Ac-ALF-LRLRLRL-ALF-NH2 we got two equilibrated structures. In the first structure we have hydrogen bonds alternating between Glutamine and Leucine creating strong interactions between the individual chains (Fig 6). The Hydrophobic residues created prevent water from getting in creating a hydrophobic core. This resulted in a very twisted peptide aggregation which can be seen in the equilibrated structure (Fig. 5). The second structure has hydrogen bonds between Leucine and Arginine. Compared to the first structure, this is more twisted most likely due to the smaller size of Arginine. This also increased interaction between the chain.
Conclusions:
The equilibrated structures created after molecular dynamics created strong hydrogen bonds due to the alternating hydrophobic-hydrophilic sequence that we used. There was also a twist created due to the hydrophobic amino acid Leucine. This prevents the non-hydrophobic amino acids from creating hydrogen bonds with water molecules.
Future Goal:
The extension of twisting between the neighboring chains differ for both structures. However to determine the differences, further structural studies must be done. Additionally the Influence of sequencing structures, as used can be seen in the structures used in this project, needs to be further studied to analyze its affect on catalytic activity. In the peptide chains, we used the artificial amino acid ALF instead of histidine. This ALF is capable of binding with metals due to the nitrogen on the para position in the its benzene ring. Creating a equilibrated metal bounded peptide structure will be the next step with this research. Creating active cites with different metals are an important facet to this project. However the way metals affect the
overall aggregation and activity of the peptide chain is to be further explored.
Acknowledgments:
The research was supported by the University of Miami Department of Chemistry in Dr. Rajeev Prabhakar’s laboratory with coordination with the American Chemical Society.
References:
- D’Souza, A., Marshall, L. R., Yoon, J. H., K u l e s h a , A . , E d i r i s i n g h e , D . I . U . , Chandrasekaran, S., Rathee, P., Prabhakar, R., &18 Page 5 Makhlynets, O. V. (2022). Peptide hydrogel with self-healing and redox-responsive properties. Nano Convergence, 9(1). doi.org/10.1186/s40580-022-00309-7
- D’Souza, A., Yoon, J. H., Beaman, H., Gosavi, P. M., Lengyel-Zhand, Z., Sternisha, A., Centola, G., Marshall, L. R., Wehrman, M. D., Schultz, K. M., Monroe, M. B. B., & Makhlynets, O. V. (2020). Nine-Residue peptide Self-Assembles in the presence of silver to produce a Self-Healing, cytocompatible, antimicrobial hydrogel. ACS Applied Materials & Interfaces, 12(14), 17091–17099. doi.org/10.1021/acsami.0c01154
- Marshall, L. R., Jayachandran, M., Lengyel-Zhand, Z., Rufo, C. M., Kriews, A., Kim, M. C.,& Korendovych, I. V. (2020). Synergistic interactions are prevalent in catalytic amyloids. ChemBioChem, 21(18), 2611–2614. doi.org/10.1002/cbic.202000205
- Song, R., Wu, X., Xue, B., Yang, Y., Huang, W., Zeng, G., Wang, J., Li, W., Cao, Y., Wang, W., Lu, J., & Dong, H. (2018). Principles governing catalytic activity of Self-Assembled short peptides. Journal of the American Chemical Society, 141(1), 223–231. doi.org/10.1021/jacs.8b08893
- Zozulia, O., Dolan, M. A., & Korendovych, I.V. (2018). Catalytic peptide assemblies. Chemical
Society Reviews, 47(10), 3621–3639.doi.org/10.1039/c8cs00080h








