Abstract
Peptides can serve a multitude of purposes in the natural world depending on the amino acid sequences they contain, length of peptides, and other characteristics. In this paper, we examine the structural differences of peptides with differing 7 residue sequences. We focused on peptides which contain alternating apolar-polar sequences versus peptides which do not. Using Molecular Dynamics (MD) simulations, we observed variations in the structure amongst the peptides used.
Introduction
Peptides can serve a multitude of purposes in the natural world depending on the amino acid sequences they contain, length of peptides, and other characteristics. Short peptides have been shown to have significant catalytic activity. [4] Furthermore, shorter peptides with alternating apolar and polar residues are more likely to self-assemble into beta sheets than larger peptides.[4] In this project, we will investigate and compare two peptides with seven alternating apolar and polar sequences (LKLKLQL and LHLHKL) and two peptides that do not exhibit this pattern (GNNQQNY and KLVFFAL). GNNQQNY was used for its catalytic efficiency in conjunction with metal like Zinc.[1] KLVFFAL was used since fragments the amyloid beta protein such as FFKLVFF and LVFF have been shown to significantly increase catalytic activity. [1 – 3] LKLKLQL has been shown to be a beta sheet forming minimalist and an efficient catalyst. [1, 4] Lastly, LHLHLKL was used since it contains histamine in two positions [2 & 4]. Histamine, in conjunction with metals such as Zinc, has shown exceptional catalytic potential. [1,4]
Computational Details
All Molecular Dynamics (MD) simulations were conducted using GROMACS. The force field used was AMBER14sb. MD simulations for all sequences of peptides were done at 100 ns. Energy was minimized on all simulations to ensure the stability of each structure. Natural volume and temperature conditions were used for all simulations. To convert the resulting XTC file from MD, we used the g cluster technique to convert to a final PDB file which contained the resulting structure of the simulations as shown in figures (1,2,3, 4, 5, and 6).
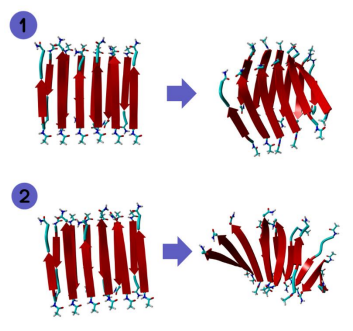
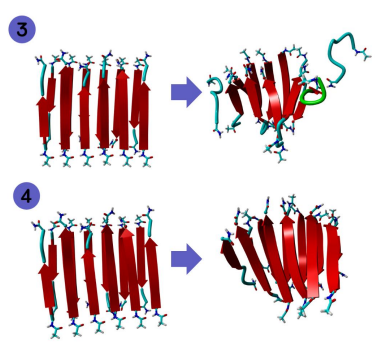
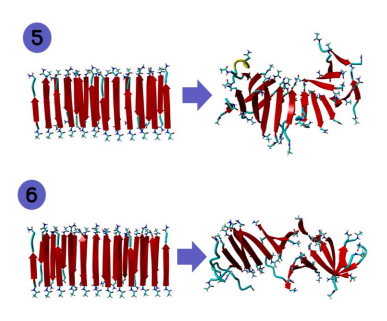
Figure 1: 6 by 2 structure of GNNQQNY before and after MD.
Figure 2: 6 by 2 structure of KLVFFAL before and after MD.
Figure 3: 6 by 2 structure of LKLKLQL before and after MD.
Figure 4: 6 by 2 structure of LHLHLKL before and after MD.
Figure 5: 12 by 2 structure of KLVFFAL before and after MD.
Figure 6: 12 by 2 structure of LKLKLQL before and after MD.
Results and Discussion
Peptides with alternating apolar and polar sequences exhibit differing degrees of twisting when compared to peptides with non-alternating sequences. Using figures 1, 2, 3, and 4, we can compare the degree of twisting for each sequence of peptides.
Non-alternating Peptides
Figures 1 and 2 contain 6 by 2 structures of the sequences GNNQQNY and KLVFFAL. We can see that KLVFFAL exhibits more twisting than GNNQQNY which is in line with the conclusions of previous research.
[1-3]
Alternating Peptides
Figures 3 and 4 contain 6 by 2 structures of the sequences LKLKLQL and LHLHKL. We can see that LKLKLQL
exhibits far more twisting than LHLHKL. Overall, the sequences that exhibited the most twisting was KLVFFAL and LKLKLQL. Increased twisting is important because it can show increased catalytical and/or physical properties of a peptide. To further examine the structural differences between these sequences, 12 by 2 structures of these sequences were run through MD in addition to the 6 by 2 structures.
Further Analysis
Figures 5 and 6 contain these 12 by 2 structures (KLVFFAL and LKLKLQL, respectively). We can see that figure five’s twisting is much more defined than that of figure six’s. However, figure six is slightly more twisted than figure five. We can attribute this difference to the alternating apolar-polar sequence of figure 6 (LKLKLQL).[1]
Conclusion
The use of peptides with alternating apolar-polar residue sequences results in structures with more twisting. The present study conducted only preliminary research on these sequences due to time constraints. The next possible step in this research would be to examine the catalytic properties of each peptide. This can be done by adding metals such as Zn2+. Future research can also examine physical properties such as elasticity and toughness.
Acknowledgements
We would like to thank the Chemistry department at the University of Miami and Professor
Rajeev Prabhakar for letting me use their lab to conduct this research. Additionally, we would
like to thank the American Chemical Society for their Project SEED summer program which has
facilitated students’ further learning of chemistry.
References
(1) Hamley, I. W. (2021). Biocatalysts based on peptide and peptide conjugate nanostructures. Biomacromolecules, 22(5), 1835-1855.
(2) Maeda, Y., Fang, J., Ikezoe, Y., Pike, D. H., Nanda, V., & Matsui, H. (2016). Molecular self-assembly strategy for generating catalytic hybrid polypeptides. PLoS One, 11(4), e0153700.
(3) Omosun, T. O., Hsieh, M. C., Childers, W. S., Das, D., Mehta, A. K., Anthony, N. R., ... & Lynn, D. G. (2017). Catalytic diversity in self-propagating peptide assemblies. Nature chemistry, 9(8), 805-809.
(4) Rufo, C. M., Moroz, Y. S., Moroz, O. V., Stöhr, J., Smith, T. A., Hu, X., ... & Korendovych, I. V. (2014). Short peptides self-assemble to produce catalytic amyloids. Nature chemistry, 6(4), 303-309.
(5) Wong, Y. M., Masunaga, H., Chuah, J. A., Sudesh, K., & Numata, K. (2016). Enzymemimic peptide assembly to achieve amidolytic activity. Biomacromolecules, 17(10), 3375-3385.






