Abstract
Computational chemistry techniques were implemented to understand one of the functions of peptides self-assemble and short 5-9amino acidss peptide chains, with various structural and catalytic properties. In the last few years, these amyloids have been used to catalyze various reactions such as hydrolysis and oxidation. They provide intrinsic details regarding the functioning of natural enzymes and help us design their synthetic analogs.
Introduction
The purpose of this project was to investigate the interactions of heterogeneous amyloids that were formed by the Ac-IHIHIGI-NH2 peptide through environmental and biological molecules portrayed in different forms of a relevant peptide. To make the inclusion of the catalytic profile as well as different kinds of molecular interactions that give stability. Hydrogen bonding, hydrophobic packing, and zinc complexation with the negative histidine are used in the system. Therefore, computational techniques were implemented by the researcher, particularly using the programs for molecular docking and all-atom classical molecular dynamics (MD) simulations.
Experimental Details
This project experiment consisted in modeling the molecular interaction happening between the catalytic amyloids that are formed by the Ac-IHIHIQI-NH2 peptide, the substrate was docked to combine the ligand and peptide through the program named “AutoDock ( version 4.0)” The researcher used the lowest energy conformer taken from the docked complex that was then used to elaborate the molecular dynamics simulations ( MD) on the amyloid-substrate complex inside an aqueous solution. The second step of the experiment consisted in structuring the Ac-IHIHIQI-NH2 molecule combined with the ligand and implementing energy minimization for 2000 steps. It was required to create a topology file and addition of water inside the box for the hydrolytic reaction. The addition of ions was needed as well to then move forward with the energy minimization before preparing the system for (MD). The final result of these 3 minimalizations of volume, pressure, and temperature produced a structure for the MD simulations.
Results & Discussion
After 2 months of research to identify the development of catalytic peptides Ac-IHIHIGI-NH2 for hydrolytic reactions, the researcher assumed that the simulations provided by the “GROMACS” and “YASARA” software applications, gave critical insight into observing and analyzing the development of synthetic analogs of the natural enzyme that can be used for future biotechnology applications. The final shape of the structure was tilted in a more horizontal way, the longer the structure, the more shifted the peptide became. In figure 1 (a), here we can observe the peptide molecule structure before going under MD simulations. On the other hand, in figure 2 (b), here we can observe the peptide structural change after going under MD simulations, we can also observe how as well the Zincs maintained a constant organization. In figure 2 (a) we can observe the root mean square deviation ( RMSD) of the backbone group complex derived from the MD trajectories. In figure 2 (b) we can view the RMSD of the protein group complex from the MD trajectories.
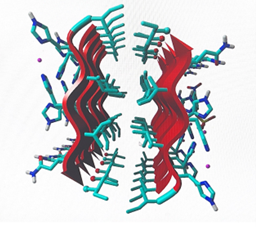
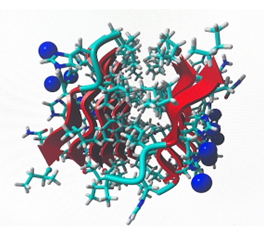
Figure 1: (a) left; (b) right.
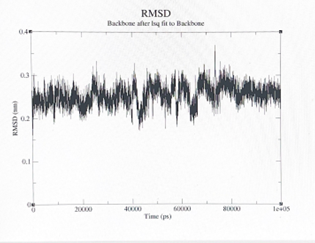
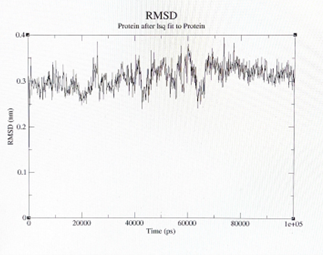
Figure 2: (a) left; (b) right.
Conclusion
In conclusion, this project has demonstrated that catalytic amyloids tend to have a high ability for synthetic reactions. This helps explain why peptides have a great ability to synthesize as well. This can be observed in all of the MD simulations elaborated in this research project were performed by using the software programs “GROMACS” and “YASARA”, where they provided visual interpretations of the molecules formed. The final structure for the peptide showed a shift in the program, compared to the first structure given.
Acknowledgements
This work was supported by the America Chemical Society, and the University of Miami Department of Chemistry. I would like to thank Professor Prabhakar, Rajeev for guiding and supporting the process of learning computational chemistry.
References
- Myungwoon Lee, Tuo Wang, Olga V. Makhlyne, Yibing Wu, Nicholas F. Polizzi, Haifan Wu, Pallavi M. Gosavi, Jan Stöhr, Ivan V. Korendovych, William F. DeGrado and Mei Hong (2017) Zinc-binding structure of catalytic amyloid from solid-state NMR.
- Liam R Marshall, Megha Jayachandran, Zsofia Lengyel-Zhand, Caroline M Rufo, Austin Kriews, Min-Chul Kim, Ivan V Korendovych (2020) Synergistic Interactions Are Prevalent in Catalytic Amyloids







