Abstract. Clavanin A (ClavA) is an antimicrobial peptide (AMP) whose purpose is to eliminate various pathogens, mainly Escherichia coli (E. coli). ClavA can bind to a zinc(II) ion which will increase its antimicrobial activity. ClavA can also be mutated to obtain different properties, mostly negative, which will affect its overall performance when eliminating pathogens. In this experiment, the properties of ClavA-H10A-H11A-Zn(II) were determined and analyzed.
Introduction. Antimicrobial peptides (AMPs) are microbial proteins that live within living organisms to kill foreign pathogens through fatally interacting with the pathogens’ DNA or creating pores in their cellular membrane [1-4]. An AMP that does this is ClavA (VFQFLGKIIHHVGNFVHGFSHVF-NH2). ClavA is a cationic, alpha-helical, amphipathic 23-amino acid peptide that targets various pathogens, most importantly Escherichia coli. ClavA can also bond with metallic ions to form a metalloAMP. When ClavA binds with a zinc(II), it causes an increase in antimicrobial activity [4-6]. Like all proteins, ClavA can be mutated. These mutations can either aid the protein or weaken its performance [4]. In this work, the properties of ClavA-H10A-H11A-Zn(II) when interacting with E. coli’s DNA will be analyzed.
Experimental Details. The ClavA structure was obtained from the Protein Data Bank. YASARA [7] was used to add Zn(II) to the protein and mutate ClavA-Zn(II). The DNA structure was constructed using Avogadro [8]. HDOCK Server [9] was used to dock the DNA and metalloAMP to create a structure for MD simulation that uses GROMACS [10]. CHARMM36 [11] force field was used to create a 6x6x6 Å box which was then filled with SPC water molecules, 55 Na+ and, 20 Cl-. The structure was energy-minimized and ran through a MD simulation to create a 120 ns trajectory. A cluster analysis of the structure was performed in addition to other calculations. YASARA and Chimera were used for molecular visualization, structural diagrams, and analysis.
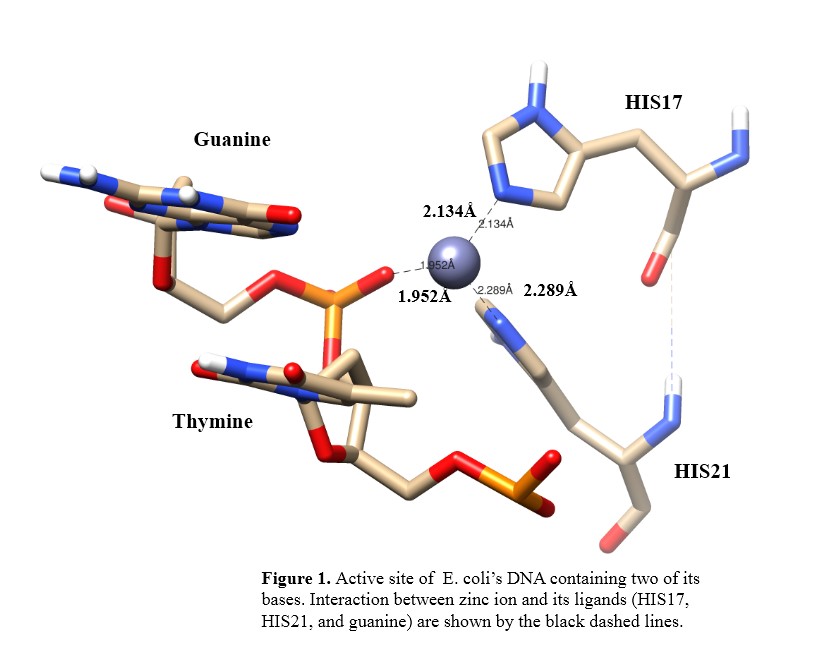
Results and Discussion. In the binding site, a zinc(II) is located next to His17, His21, and guanine. The phosphate oxygen atom from guanine and the nitrogen atoms from His17 and His21 binds the zinc ion (Figure 1).
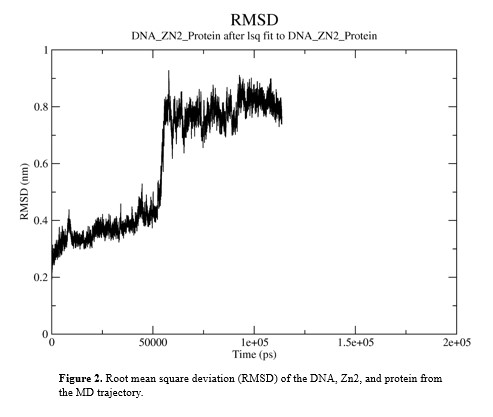
The RMSD of the structure was calculated and tt indicated a stable conformation from 60 ns to 120 ns. Within this time the RMSD value remained between 6 and 10 Å (Figure 2).
Changes did occur to the structure during the simulation. When comparing the initial structure to the cluster structure, the maximum change in distance between the zinc ion and its ligands was 0.09 Å, His17 experienced a 90 degrees rotation, and the protein rotated 45 degrees. The ends of the protein underwent high changes in position due to the protein’s terminals losing their alpha-helicity while other residues had little. After 60 ns of the structure in the simulation, its radius of gyration dramatically increased from on average 19.5 Å to 20Å due to the protein’s rotation. Also, the average number of hydrogen bonds between the protein and DNA was 3 but there was a subtle increase after 60 ns.
Conclusion. The results showed that conformational changes did occur throughout the protein and the active site. These changes consisted of the distances between the zinc ion and its ligands, the movements of the protein and its residues, the structure’s radius of gyration, and the number of hydrogen bonds between the protein and DNA. The outcomes of this study have advanced our understanding of this peptide and will lead us to gather further knowledge about ClavA.
Acknowledgements. This study was supported by the efforts and guidance of Dr. Rajeev Prabhakar, Gaurav Sharma, and Vindi Mahesha.
References.
(1) Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4.
(2)Felício, M.R.; Silva, O.N.; Gonçalves, S.; Santos, N.C.; Franco, O.L. Peptides with Dual Antimicrobial and Anticancer Activities. Front. Chem. 2017, 5, 5.
(3) Ulm, H.; Wilmes, M.; Shai, Y.; Sahl, H.-G. Antimicrobial Host Defensins—Specific Antibiotic Activities and Innate Defense Modulation. Front. Immunol. 2012, 3, 249.
(4) Samuel A. Juliano, Scott Pierce, James A. deMayo, Marcy J. Balunas, and Alfredo M. Angeles-Boza. Exploration of the Innate Immune System of Styela clava: Zn2+ Binding Enhances the Antimicrobial Activity of the Tunicate Peptide Clavanin A. Biochemistry 2017 ,56, 10, 1403-1414.
(5) Searle S. Duay, Gaurav Sharma, Rajeev Prabhakar, Alfredo M. Angeles-Boza, and Eric R. May. Molecular Dynamics Investigation into the Effect of Zinc(II) on the Structure and Membrane Interactions of the Antimicrobial Peptide Clavanin A. J. Phys. Chem. B 2019, 123, 15, 3163-3176.
(6) Lee, Tzong-Hsien, Kristopher N Hall, and Marie-Isabel Aguilar.; Antimicrobial peptide structure and mechanism of action: a focus on the role of membrane structure. Current topics in medicinal chemistry 16.1 2016: 25-39.
(7) Krieger, Elmar, Günther Koraimann, and Gert Vriend. "Increasing the precision of comparative models with YASARA NOVA—a self‐parameterizing force field.; Proteins: Structure, Function, and Bioinformatics 47.3 2002: 393-402.
(8) Hanwell, Marcus D., et al.; Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. Journal of cheminformatics 4.1 2012: 17.
(9) Yan, Yumeng, et al. " HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic acids research 2017, W365-W373.
(10) Van Der Spoel, David, et al.; GROMACS: fast, flexible, and free." Journal of computational chemistry, 2005: 1701-1718.
(11) Huang, Jing, and Alexander D. MacKerell Jr. ;CHARMM36 all‐atom additive protein force field: Validation based on comparison to NMR data; Journal of computational chemistry 2013, 2135-2145.





