Abstract
Amino acids are chemical compounds that each serve a respective purpose in an environment. All amino acids, or residues, have a central carbon atom connected to a hydrogen, carboxyl group, an amine group, and an R group.1 Think of each amino acid as a different color, each color with its own respective elementary function. Then, visualize peptides as unique combinations of colors, each combination yielding more advanced functions such as building muscle or making hormones.2 It is of importance to study the arrangements of a peptide sequence to potentially find environmentally effective solutions such as biocatalysts.4
Introduction
Peptides and amino acids are truly a fundamental part of the molecular world, as enzymes and proteins heavily depend on the fruits from amino acid sequencing. Amino acids can “connect” or bind together by means of a dehydration reaction, forming an aggregation.3 As the complexity of the aggregation increases, its function, structure, and versatility also rises in intricacy.5 This research will focus primarily on the secondary structure of amyloids, or peptide clusters between 2-7 amino acids.
There are 7 peptides that will be used in this research: lysine (K), leucine (L), glutamine (G), valine (V), histidine (H), arginine (R), asparagine (N), and tyrosine (Y). The peptides in this research are composed of 7 amino acids, and the examined report included the use of four different amino sequences; GNNQQNY, KLVFFAL, LKLKLQL, LHLHLRL. LKLKLQL and LHLHLRL were chosen to observe the relationship between alternating amino acid sequencing and a secondary structure called a β-sheet.6 The sequences KLVFFAL and GNNQQNY were also chosen to observe specific self-assembling patterns.7
Experimental Details
To begin constructing the peptide sequences, I used YASARA to visualize and build each peptide aggregate. The structures were then ran with force field Amber14 using Gromacs through a technique called md. After running the topology with the solvent, each peptide structure was then energy minimized and ran with G-cluster.
Results and Discussion

Figure 1 The diagram shows input peptide structures on the left and shows output peptide structures on the right. “A” represents sequence GNNQQNY, and “B” represents sequence LKLKLQL, “C” represents sequence “LHLHLRL”, and “D” represents sequence “KLVFFAL”
As shown in Figure 1, the output peptide structures for B structures appear to be more helical than A structures due to the alternating peptide sequence in B structures. What further differentiates both A and B is the apparent entropy of each structure, as B also resembles a more sporadic or flexible nature in each peptide.
Output structures C and D introduce expanse or a sense of stretch to peptide structures. To elaborate, output structure D appears to not only be helical, but also have a bend on peptide ends. Output structure C exhibits this bend but in less intensity. Both output structures have an equal helical nature, however, both structures appear to be moving in opposite helical directions.
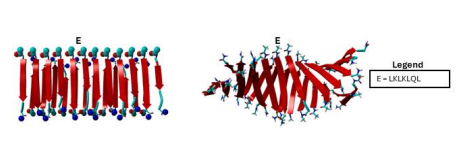
Figure 2 The diagram shows input peptide structures on the left and shows output peptide structures on the right. “E” represents sequence “LKLKLQL”.
Figure 2 highlights the advantages of having a 12 by 2 structure in opposition to a 6 by 2 structure, as we can see more of the helix generated by each peptide sequence. Output structure E appears to complete output structure B, and we can further denote the direction of the helix as more structure is shown.
Output structure B seems to have the most intense β-sheets in comparison to all output structures. All input structures from each Figure seem similar, if not identical. Figure 3 The diagram shows input peptide structures on the left and shows output peptide structures on the right. “E” represents sequence “LKLKLQL”.
Conclusion
Higher number peptide sequences are needed to further examine the self-assembling properties of output structures. Additional studies could be to examine the metallocatalytic properties of histidine, measure physical toughness, or observe beta sheet formation in other solvents.
Acknowledgements
Thank you to Project SEED for offering me the opportunity to engage in such captivating research. Special thanks to the University of Miami’s Department of Chemistry and Professor Prabhakar for making this program the best summer I have had.
References
(1) Ahern , K., Rajagopal, I., & Tan, T. (2018). Biochemistry Free For All (Ahern, Rajagopal, and Tan). Oregon State University.
(2) Cleveland Clinic. (2024). Amino Acids. Amino Acids.
(3) Forbes, J., & Krishnamurthy, K. (2024). Biochemistry, Peptide. StatPearls.
(4) Hamley, I. W. (2021). Biocatalysts based on peptide and peptide conjugate nanostructures. Biomacromolecules, 22(5), 1835-1855.
(5) Rogers, K. (2016, August 3). What Is the Difference Between a Peptide and a Protein?. Encyclopedia Britannica. https://www.britannica.com/story/what-is-the-differencebetween-a-peptide-and-a-protein.
(6) Rufo, C. M., Moroz, Y. S., Moroz, O. V., Stöhr, J., Smith, T. A., Hu, X., ... & Korendovych, I. V. (2014). Short peptides self-assemble to produce catalytic amyloids. Nature chemistry, 6(4), 303-309.
(7) Wong, Y. M., Masunaga, H., Chuah, J. A., Sudesh, K., & Numata, K. (2016). Enzyme-mimic peptide assembly to achieve amidolytic activity. Biomacromolecules, 17(10), 3375-3385.





