Abstract
Peptides are short chains of amino acids which play crucial roles in many biological processes, including hormone production, immune function, and cell signaling. Peptides are also known to be able to perform self-aggregation which is driven by non-covalent interactions between peptide molecules, such as hydrogen bonding, hydrophobic interactions, and ionic interactions. These interactions can lead to a more diverse structure which is the aim to demonstrate in this study.
Introduction
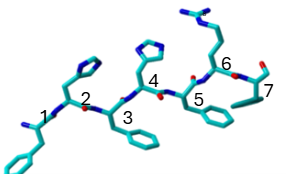
In this study, we designed and studied seven-residue peptides with a conserved Phenylalanine-Histidine repeating in the first five positions. We chose F-H because Phenylalanine, is known to drive hydrophobic interactions and pi-pi stacking, both critical for initiating self-assembly. Histidine, on the other hand, introduces a unique pH-sensitive component, potentially influencing aggregation behavior in varying environments. The key variation lies at the sixth residue, which alternates between Arginine (R), a positively charged amino acid, and Aspartate (D), a negatively charged amino acid. Both variants terminate with Phenylalanine. This controlled substitution allows us to examine how terminal charge differences influence self-aggregation behavior.
Computational Details
All of the Molecular Dynamics simulations were conducted using a molecular dynamic open-source program called GROMACS. The force field that we used to conduct this study was CHARMM36. We minimized the energy on all simulations to ensure the stability of each structure. We enclosed the protein within a GROMACS simulation box in which we added our solvent as well as ions to neutralize the solvent and to maintain similar conditions as blood. All temperature, volume, and pressure all simulate natural conditions. Using the G-cluster tool, the resulting XTC file from our MD simulations was converted to a final PDB file, which contained the final structure of our simulations.
Results and discussion
When comparing the input file of the 6x2 as well as the 12x2 to the output file it will be shown that the outputs have more of a spiral form rather than the original linear. Our molecular dynamics simulations revealed that these peptides, initially synthesized as linear amino acid sequences, underwent structural transformations.
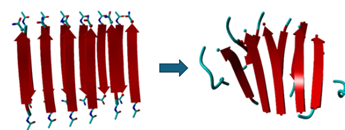
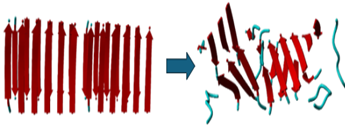
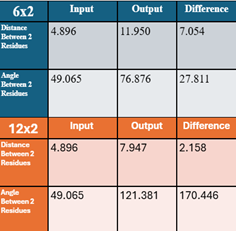
As seen in figures 1 and 2, the structure hasnow a twisted structure. Crucially, our analysis in figure 3 shows significant alterations in the inter-residue distances and bond angles within the peptide. Our results indicate these transformations are directly linked to the peptide's ability to self-assemble into structures and significantly influence its catalytical and physical properties.
Conclusion
Ultimately, peptides with alternating charges showed distinct structural differences compared to their initial configurations. Specifically, the outputs showed more spirals and twisting, along with greater distances and angles between two specific residues. It's important to note that the current study conducted only initial research on these sequences, as comprehensive analysis was limited by time.
Acknowledgements
I wish to express gratitude to Professor Rajeev Prabhaker and the Chemistry department at the University of Miami for letting me use their lab to conduct this research. As well as the American Chemical Society for allowing me the opportunity to be in their Project SEED summer program.
References
Abraham, Mark James, et al. “GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers.” SoftwareX, vol. 1-2, Sept. 2015, pp. 19–25, www.sciencedirect.com/science/article/pii/S2352711015000059, https://doi.org/10.1016/j.softx.2015.06.001
Ho, Bosco, and Ken A Dill. “Folding Very Short Peptides Using Molecular Dynamics.” PLOS Computational Biology, vol. 2, no. 4, 14 Apr. 2006, pp. e27–e27, www.ncbi.nlm.nih.gov/pmc/articles/PMC1435986/, https://doi.org/10.1371/journal.pcbi.0020027.
Lee, Sungeun, et al. “Self-Assembling Peptides and Their Application in the Treatment of Diseases.” International Journal of Molecular Sciences, vol. 20, no. 23, 21 Nov. 2019, www.ncbi.nlm.nih.gov/pmc/articles/PMC6928719/, https://doi.org/10.3390/ijms20235850.







